Note
Go to the end to download the full example code.
Raypaths in layered and gradient models#
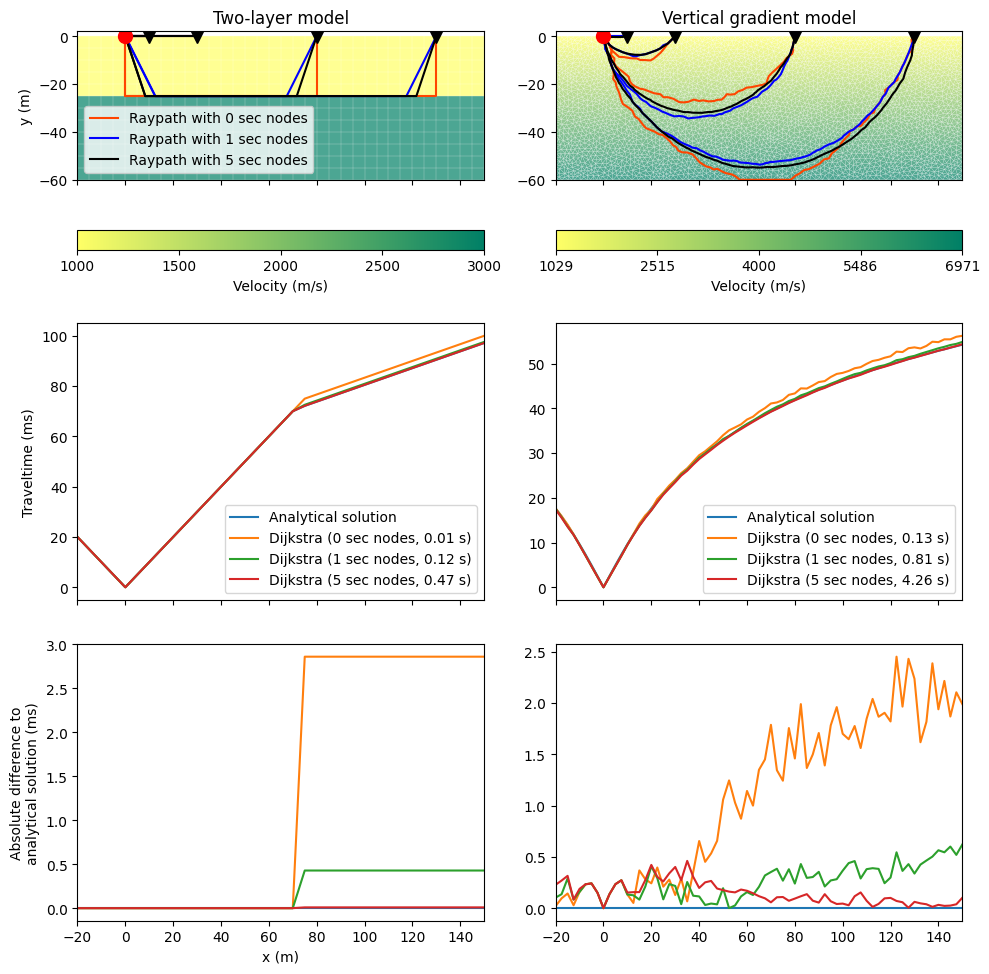
This example performs raytracing for a two-layer and a vertical gradient model and compares the resulting traveltimes to existing analytical solutions. An approximation of the raypath is found by finding the shortest-path through a grid of nodes. The possible angular coverage is small when only corner points of a cell (primary nodes) are used for this purpose. The angular coverage, and hence the numerical accuracy of traveltime calculations, can be significantly improved by a few secondary nodes along the cell edges. Details can be found in Giroux & Larouche (2013).
Two-layer model#
We start by building a regular grid.
mesh_layered = mt.createGrid(
np.arange(-20, 155, step=5, dtype=float), np.linspace(-60, 0, 13))
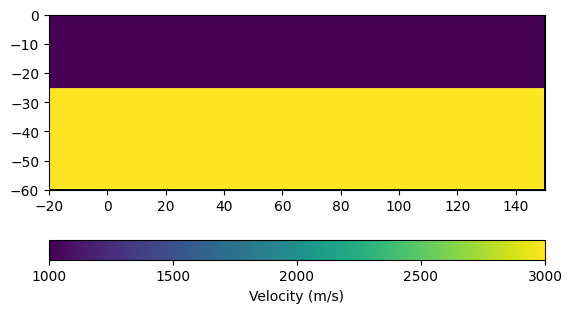
We now construct the velocity vector for the two-layer case by iterating over the cells. Cells above 25 m depth are assigned \(v = 1000\) m/s and cells below are assigned \(v = 3000\) m/s.
vel_layered = np.zeros(mesh_layered.cellCount())
for cell in mesh_layered.cells():
if cell.center().y() < -25:
vel = 3000.0
else:
vel = 1000.0
vel_layered[cell.id()] = vel
ax, cb = pg.show(mesh_layered, vel_layered, label="Velocity (m/s)")
We now define the analytical solution. The traveltime at a given offset x is the minimum of the direct and critically refracted wave, where the latter is governed by Snell’s law.
def analyticalSolution2Layer(x, zlay=25, v1=1000, v2=3000):
"""Analytical solution for 2 layer case."""
tdirect = np.abs(x) / v1 # direct wave
alfa = asin(v1 / v2) # critically refracted wave angle
xreflec = tan(alfa) * zlay * 2. # first critically refracted
trefrac = (x - xreflec) / v2 + xreflec * v2 / v1**2
return np.minimum(tdirect, trefrac)
Vertical gradient model#
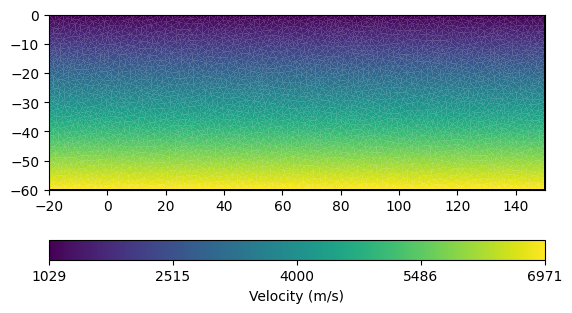
We first create an unstructured mesh:
A vertical gradient model, i.e. \(v(z) = a + bz\), is defined per cell.
The traveltime for a gradient velocity model is given by:
The loop below calculates the travel times and makes the comparison plot.
fig, ax = plt.subplots(3, 2, figsize=(10, 10), sharex=True)
for j, (case, mesh, vel) in enumerate(zip(["layered", "gradient"],
[mesh_layered, mesh_gradient],
[vel_layered, vel_gradient])):
pg.boxprint(case)
if case == "gradient":
ana = analyticalSolutionGradient
elif case == "layered":
ana = analyticalSolution2Layer
for boundary in mesh.boundaries():
boundary.setMarker(0)
xmin, xmax = mesh.xmin(), mesh.xmax()
mesh.createNeighborInfos()
# In order to use the Dijkstra, we extract the surface positions >0
mx = pg.x(mesh)
my = pg.y(mesh)
px = np.sort(mx[my == 0.0])
# A data container with index arrays named s (shot) and g (geophones) is
# created and filled with the positions and shot/geophone indices.
data = pg.DataContainer()
data.registerSensorIndex('s')
data.registerSensorIndex('g')
for i, pxi in enumerate(px):
data.createSensor([pxi, 0.0])
if pxi == 0.0:
source = i
nData = len(px)
data.resize(nData)
data['s'] = [source] * nData # only one shot at first sensor
data['g'] = range(nData) # and all sensors are receiver geophones
# Draw initial mesh with velocity distribution
pg.show(mesh, vel, ax=ax[0, j], label="Velocity (m/s)", hold=True,
logScale=False, cMap="summer_r", coverage=0.7)
drawMesh(ax[0, j], mesh, color="white", lw=0.21)
# We compare the accuracy for 0-5 secondary nodes
sec_nodes = [0, 1, 5]
t_all = []
durations = []
paths = []
mgr = TravelTimeManager()
cols = ["orangered", "blue", "black"]
recs = [1, 3, 8, 13]
for i, n in enumerate(sec_nodes):
# Perform traveltime calculations and log time with pg.tic() & pg.toc()
pg.tic()
res = mgr.simulate(vel=vel, scheme=data, mesh=mesh, secNodes=n)
# We need to copy res['t'] here because res['t'] is a reference to
# an array in res, and res will be removed in the next iteration.
# Unfortunately, we don't have any reverence counting for core objects yet.
t_all.append(res['t'].array())
durations.append(pg.dur())
pg.toc("Raytracing with %d secondary nodes:" % n)
for r, p in enumerate(recs):
if r == 0:
lab = "Raypath with %d sec nodes" % n
else:
lab = None
recNode = mgr.fop.mesh().findNearestNode([sensors[p], 0.0])
sourceNode = mgr.fop.mesh().findNearestNode([0.0, 0.0])
path = mgr.fop.dijkstra.shortestPath(sourceNode, recNode)
points = mgr.fop.mesh().positions(withSecNodes=True)[path].array()
ax[0, j].plot(points[:,0], points[:,1], cols[i], label=lab)
t_ana = ana(px)
# Upper subplot
ax[1, j].plot(px, t_ana * 1000, label="Analytical solution")
for i, n in enumerate(sec_nodes):
ax[1, j].plot(px, t_all[i] * 1000,
label="Dijkstra (%d sec nodes, %.2f s)" % (n, durations[i]))
ax[2, j].plot(px, np.zeros_like(px), label="Zero line") # to keep color cycle
for i, n in enumerate(sec_nodes):
ax[2, j].plot(px, np.abs(t_all[i] - t_ana) * 1000)
ax[1, j].legend()
# Draw sensor positions for the selected receivers
for p in recs:
ax[0, j].plot(sensors[p], 0.0, "kv", ms=10)
ax[0, j].plot(0.0, 0.0, "ro", ms=10)
ax[0, j].set_ylim(mesh.ymin(), 2)
ax[0, 0].set_title("Two-layer model")
ax[0, 1].set_title("Vertical gradient model")
ax[0, 0].legend()
ax[0, 0].set_ylabel("y (m)")
ax[1, 0].set_ylabel("Traveltime (ms)")
ax[2, 0].set_ylabel("Absolute difference to\nanalytical solution (ms)")
ax[2, 0].set_xlabel("x (m)")
fig.tight_layout()
################################################################################
# layered #
################################################################################
Raytracing with 0 secondary nodes: Elapsed time is 0.00 seconds.
Raytracing with 1 secondary nodes: Elapsed time is 0.02 seconds.
Raytracing with 5 secondary nodes: Elapsed time is 0.12 seconds.
################################################################################
# gradient #
################################################################################
Raytracing with 0 secondary nodes: Elapsed time is 0.04 seconds.
Raytracing with 1 secondary nodes: Elapsed time is 0.17 seconds.
Raytracing with 5 secondary nodes: Elapsed time is 1.07 seconds.